DNA elimination in parasitic nematodes
In the human and pig parasitic nematode Ascaris, DNA elimination occurs during early embryonic development, at the 4- to 16-cell stage, with DNA loss occurring in five independent pre-somatic cells (Fig. 1A). Through genome sequencing and comparison, we identified ~1,000 germline-expressed genes that are eliminated in Ascaris. Further analysis of DNA elimination in two additional parasitic nematodes, Parascaris from horses and Toxocara from dogs, revealed that ~1,000-2,000 genes are eliminated, respectively. About 35% of these eliminated genes are conserved among these nematodes and are mainly expressed in the testis. Our data suggests that DNA elimination may serve as a mechanism of gene regulation – rather than repressing these germline-specific genes in somatic cells, these genes are removed from the genome as an irreversible mechanism of gene silencing.
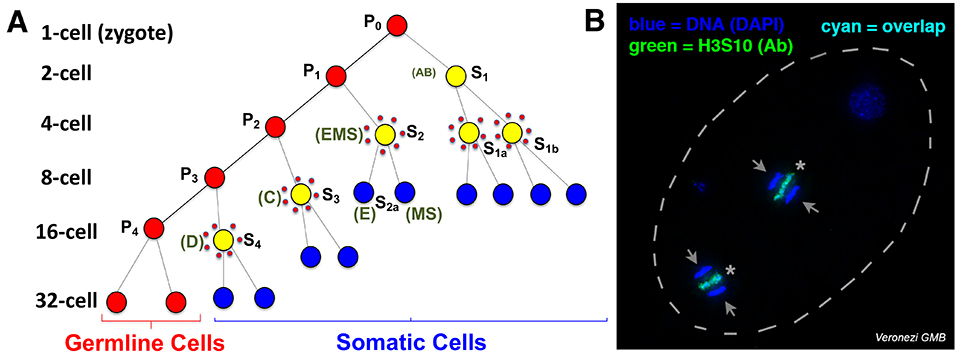
Fig. 1. Programmed DNA elimination in Ascaris. A. Cell lineages showing DNA elimination in five pre-somatic cells (yellow surrounded by red dots). B. H3S10P staining of DNA being eliminated simultaneously in two cells of the 4-cell embryo. Arrows: retained DNA; *: eliminated DNA.
We are beginning to unveil the mechanisms of Ascaris DNA elimination. We found that chromosomal breaks lead to all chromosome ends removal and remodeling, as well as an increase in the number of chromosomes from 24 to 36. We also discovered that centromere deposition changes (the lack of centromere in to-be-eliminated DNA in holocentric chromosomes) that facilitate DNA loss during mitosis. Our current data suggests that a non-sequence-specific mechanism is involved in Ascaris chromosome breaks, with the break regions becoming more chromatin accessible right before DNA elimination mitosis. Efforts are underway to address what mechanisms might be involved in the recognition of the break regions and the generation of the breaks.
DNA elimination in free-living nematodes
Oscheius tipulae is a free-living nematode that is ubiquitous around the world. It has been used as a comparative model to C. elegans to analyze the evolution of developmental pathways. Recently, O. tipulae was suggested to eliminate DNA. Using staged embryos and DNA FISH, we show that O. tipulae DNA elimination occurs during embryogenesis at the 8-16 cell stages (Fig. 2). We identified a conserved motif, named Sequence For Elimination (SFE), at the junctions of retained and eliminated regions. The SFE sequence is necessary for DNA elimination. We also revealed many alternative SFEs at the chromosome ends, likely providing flexibility in the sequences eliminated and a fail-safe mechanism for DNA elimination. These studies establish O. tipulae as a genetic and functional model to study DNA elimination in a metazoan.
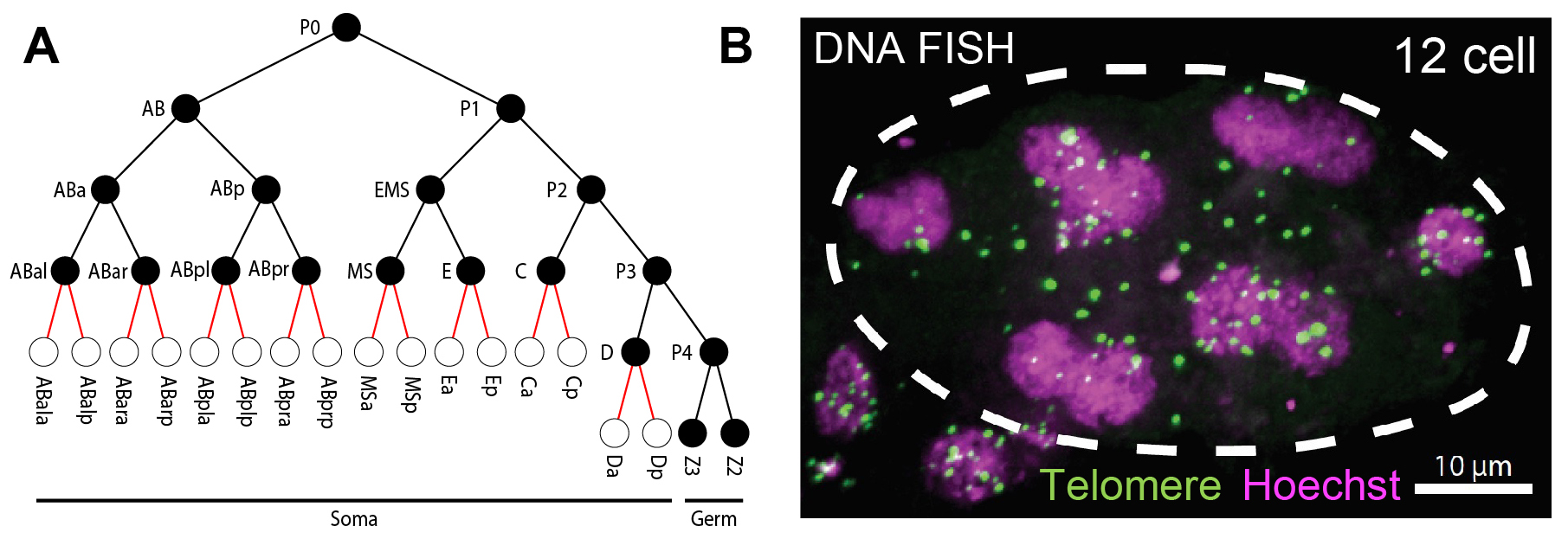
Fig. 2. Programmed DNA elimination in O. tipulae. A. Cell lineages showing DNA elimination occurs between 8 to 16 cells. B. Telomere FISH (green) and DNA Hoechst staining (magenta) in a 12-cell O. tipulae embryo. Note many of the FISH-labeled telomeric sequences are not in the nuclei, suggesting their removal from the ends of the chromosomes.
O. tipulae is similar to the model organism C. elegans in their physiology, life cycle, lab maintenance, and ability to carry out genetic manipulation. The small genome (60 Mb), complete telomere-to-telomere genome assembly and comprehensive transcriptomes, rapid generation time, transparent body and ease of microscopic analyses, and the ability to culture large quantities for biochemical and molecular studies makes O. tipulae an ideal lab model. We are carrying out in-depth analyses on the functions, mechanisms, and evolution of DNA elimination in O. tipulae and its related species.
DNA elimination in copepods
In the freshwater copepod Mesocyclops edax, DNA elimination occurs during the 16-32 cell stages (Fig. 3) with an estimated 80-90% of the germline genome eliminated to form the somatic genome. Unlike the increase in somatic chromosome number following DNA elimination in the parasitic nematodes, copepod chromosome number does not change after DNA elimination.
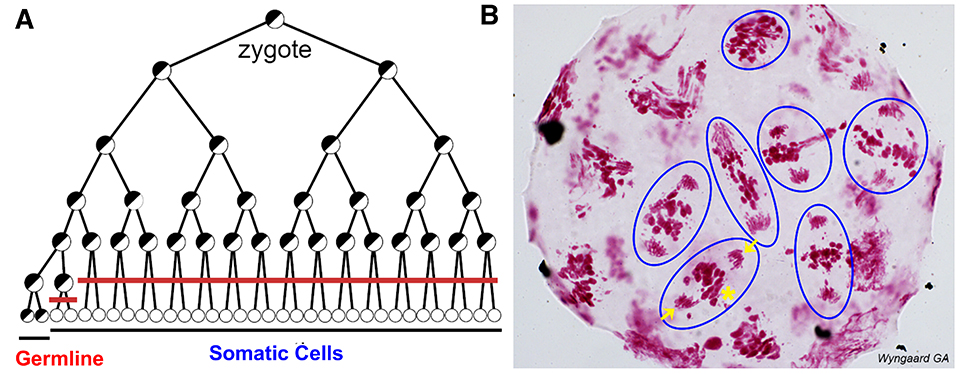
Fig. 3. Programmed DNA elimination in copepod M. edax. A. Cell lineages showing DNA elimination in 16 pre-somatic cells (red line). B. Feulgen DNA staining in a 16-32-cell M. edax embryo; blue circles illustrate cells undergoing a DNA elimination mitosis. Arrows: retained DNA; *: eliminated DNA (only one exemplary cell is marked).
We hypothesize that copepod DNA elimination likely involves DNA looping, break, and chromosome fusion. To examine this hypothesis, we are sequencing and compare the 7.5 Gb germline and 1.5 Gb somatic genome of M. edax using Illumina and PacBio. We will identify the retained and eliminated DNA and the sites where DNA breaks. The results will determine if there are common features at the DNA breaks that would suggest a potential mechanism for recognizing and making the breaks. We will also examine circular DNA during elimination as potential intermediates for the looping and fusion model of DNA elimination. In addition, we are using the copepod culture in the lab to develop and carry out functional and molecular analysis on copepod DNA elimination.